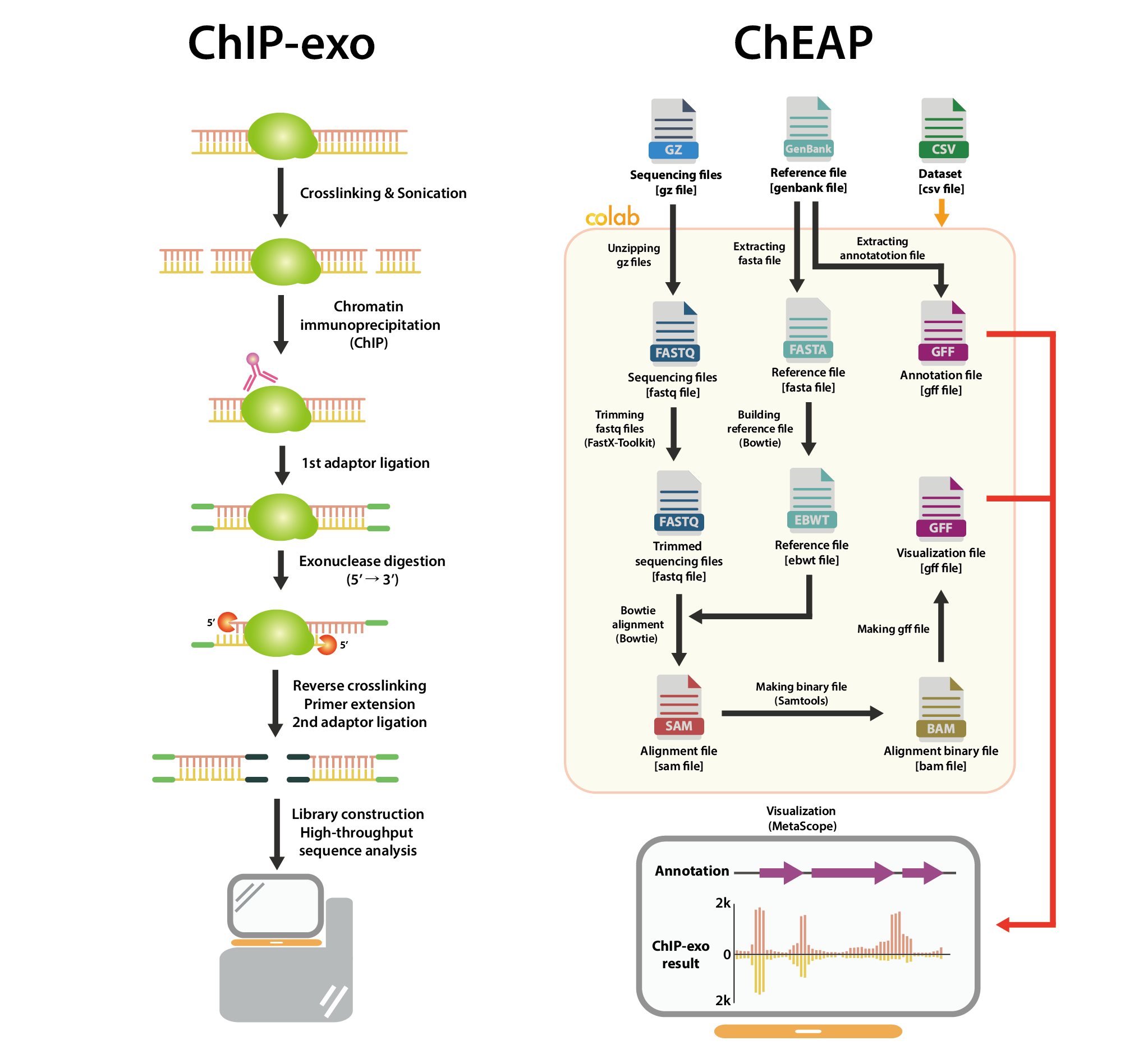
ChIP-exo Analysis Pipeline (ChEAP) that executes the one-step process, starting from trimming and aligning raw sequencing reads to visualization of ChIP-exo results. The pipeline was implemented on the interactive web-based Python development environment – Jupyter Notebook, which is compatible with the Google Colab cloud platform to facilitate the sharing of codes and collaboration among researchers. Additionally, users could exploit the free GPU and CPU resources allocated by Colab to carry out computing tasks regardless of the performance of their local machines.
[Citation information]
ChEAP: ChIP-exo Analysis Pipeline and the investigation of Escherichia coli RpoN protein-DNA interactions. Bang I#, Nong LK#, Park JY, Le HT, Lee SM, Kim D. Comput Struct Biotechnol J. 2022 Dec 02. doi:10.1016/j.csbj.2022.11.053.
Using in
- Colab or jupyter notebook : Download .ipynb file
Using Colab in local runtime : https://research.google.com/colaboratory/local-runtimes.html - Local computer : Download .py file ( python 3 )